R Using Anaconda Evnironment
Step 1: Installing R Additional Package with Anaconda
Section titled “Step 1: Installing R Additional Package with Anaconda”To install R additional packages with Anaconda, run below command in the SSH terminal (CLI), for tutorial in accessing CLI, please refer to Shell Access and Useful Command
# Load Anaconda Environment Module$ module load anaconda
# Add conda-forge Channels$ conda config --add channels conda-forge
# Create Anaconda Virtual Environment$ conda create –n <environment name> <packages to install>
# Create Anaconda Virtual Environment (Example)$ conda create -n R_test_env r-base=4.3 r-cairo r-gifi
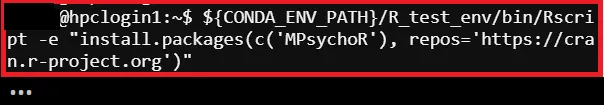
# Install Missing Package (Example)$ ${CONDA_ENV_PATH}/R_test_env/bin/Rscript -e "install.packages(c('MPsychoR'), repos='https://cran.r-project.org')"
# Purge Module$ module purge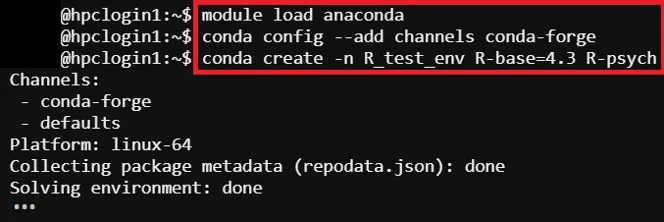

Step 2: Prepare R Program Source Code
Section titled “Step 2: Prepare R Program Source Code”Example source code path -> /home/$USER/job_template/R/MPsychoR_test.R
# Load librarieslibrary(MPsychoR)library(Gifi)library(Cairo)# Load the built-in datasetdata("zareki")
# Extract subtraction items (columns containing "subtr")zarsub <- zareki[, grep("subtr", colnames(zareki))]
# Perform nonlinear principal component analysisprinzar <- princals(zarsub)
# ---------------------------------------------------------# 1. Export the loadings plot to PNG using Cairo# ---------------------------------------------------------CairoPNG(filename = "zareki_subtraction_loadings.png", width = 800, height = 600, dpi = 150, bg = "white")
plot(prinzar, main = "Zareki Subtraction Items Loadings Plot")
dev.off()
cat("Loadings plot saved as 'zareki_subtraction_loadings.png'\n in directory:", getwd(), "\n")
# ---------------------------------------------------------# 2. Export the text results (summary) as a PNG# ---------------------------------------------------------# Capture the printed outputoutput_text <- capture.output(print(prinzar))
CairoPNG(filename = "zareki_subtraction_results.png", width = 1000, height = 1400, dpi = 150, bg = "white")
# Create an empty plot and add textplot.new()par(mar = c(2, 2, 3, 2)) # Small marginstitle(main = "Princals Results: Zareki Subtraction Items", cex.main = 1.6, font.main = 2)
# Render the captured text on the canvastext(x = 0.02, y = 0.98, labels = paste(output_text, collapse = "\n"), adj = c(0, 1), # Left- and top-aligned cex = 0.85, # Text size (adjust if needed) family = "mono") # Monospace font for clean alignment
dev.off()
cat("Text results saved as 'zareki_subtraction_results.png'\n in directory:", getwd(), "\n")Step 3: Prepare Job Template Script
Section titled “Step 3: Prepare Job Template Script”Pre-configured template script path -> /home/$USER/job_template/slurm_job/MPsychoR_test_conda.sh
#!/bin/bash#SBATCH --job-name=MPsychoR_test_conda ## Job Name#SBATCH --partition=shared_cpu ## Partition for Running Job#SBATCH --nodes=1 ## Number of Compute Node#SBATCH --ntasks-per-node=1 # Number of Task per Compute Node#SBATCH --cpus-per-task=2 ## Number of CPU per task#SBATCH --time=60:00 ## Job Time Limit (i.e. 60 Minutes)#SBATCH --mem=10GB ## Total Memory for Job#SBATCH --output=./%x%j.out ## Output File Path#SBATCH --error=./%x%j.err ## Error Log Path
## Initiate Environment Modulesource /usr/share/modules/init/profile.sh
## Reset the Environment Module componentsmodule purge
## Load Modulemodule load anaconda
## Run user command in conda environemnt## Do not use conda activate command, use absolute path instead${CONDA_ENV_PATH}/R_test_env/bin/Rscript \/home/${USER}/job_template/R/MPsychoR_test.R
## Clear Environment Module componentsmodule purgeStep 4: Create Template (Web Interface Feature)
Section titled “Step 4: Create Template (Web Interface Feature)”To submit HPC via web interface a job template is required, details please refer to: HPC Job Template For Job submission via CLI Terminal, please skip this step.
Step 5: Submit HPC Job
Section titled “Step 5: Submit HPC Job”Guides for submitting HPC job, please refer to: HPC Job Submission
Step 6: Remove user’s Conda (R) environment (Optional)
Section titled “Step 6: Remove user’s Conda (R) environment (Optional)”# Remove user’s Conda (R) environment$ rm –rf /home/$USER/.conda/envs/<environment name>
# Remove user’s Conda (R) environment (Example)$ rm –rf /home/$USER/.conda/envs/R_test_env